Regulatory mechanisms of mechanotransduction in genome instability
Review Article
Genome Instability & Disease 3, 311–316 (2022)
Abstract
During ontogeny and adult homeostasis, mechanical forces provide cells with important signals that guide cellular and tissue behavior. In addition, cells receive and transmit mechanical signals through mechanotransduction that control cell proliferation, differentiation, and death. Dysregulation of the mechanical microenvironment and the internal mechanical system of cells can lead to a variety of diseases, including tumors. Meanwhile, genomic instability is one of the key steps in cancer initiation and progression. Although numerous experiments have suggested a link between cellular mechanics and genome instability, the underlying mechanisms are unclear. Here, we discuss how the extracellular mechanical microenvironment, mechanosensing receptors, cytoskeletal systems, and mechanosignaling of nuclear mechanotransduction affect genomic instability.
Introduction
As the place where cells live, the homeostasis of their microenvironment is critical to the entire life process (Chen & Song, 2022; Gao, et al., 2020). The main point of traditional medicine research lies in chemical signals such as macromolecules (including proteins and polysaccharides), and biochemical factors (Humphrey et al., 2014; Schreiber et al., 2007). The feedback of mechanical forces is often ignored. However, proper regulation of cellular function in the body also requires the integration of numerous mechanical stimuli from the extracellular matrix (ECM), cellular contact from neighboring cells, surrounding fluids, the stiffness of the static matrix, topological properties, cell squeezing forces, and the shear stress, tension, and pressure of the fluid environment (Hayward et al., 2021). Under steady-state conditions, cells can adapt to the cost of exposure to these forces by rapidly allosteric mechanosensitive structures and cytoskeletons to develop corresponding resistances. However, when cells are suddenly subjected to biomechanical stimuli beyond their normal levels, such as excessive shear, tension, pressure, or gravity, cells can alter gene expression in response to these biomechanical changes, and in some cases even change cell types, induce transdifferentiation and cause disease or even tumor (Hayward et al., 2021). Accordingly, there is growing evidence that various mechanical signals can influence key decision-making processes, including regulation of cell proliferation, cell differentiation, and cell death (Fernandez-Sanchez et al., 2015).
Cells can sense extracellular mechanical signals in a variety of ways and translate these information into signaling events triggering biological responses, which is called mechanotransduction (Strzyz, 2016). The key cellular structures involved in mechanotransduction are cell surface receptors, including integrins in focal adhesions and cadherins in adhesion junctions, that enable cells to attach to the ECM or other cells (Dieterle et al., 2021; Sun et al., 2016). In addition, some mechanosensitive ion channels are also stimulated directly or indirectly by mechanical signals (Jin et al., 2020; Kefauver et al., 2020). They transmit mechano-mechanical signals to the myosin motor-mediated actin cytoskeletal system, inducing conformational changes in proteins and subsequent regulation of protein interactions and functions, which enable the translation of mechanical forces into biochemical signals, which are then passed through Shuttle proteins bring signals into the nucleus, or extracellular forces directly modulate nuclear shape dynamics and integrity, thereby regulating specific genetic programs, and ultimately altering their own behavior or changing their surrounding microenvironment to adapt to the corresponding mechanical force stimuli (Strzyz, 2016; Wozniak & Chen, 2009). These mechanisms are fundamental for cells to detect and respond to the mechanical properties of their microenvironment, including ECM stiffness, attachment to other cells, stretching of tissues or shear stress imposed by flowing fluids, and appropriate responses to these signals.
Maintaining the integrity and fidelity of the genome is the key to the normal proliferation and differentiation of cells, as well as the prerequisite and guarantee for the survival and reproduction of all organisms (Jeggo et al., 2016). Heritable variations in genomic DNA molecules are called genomic instability. This is a persistent sequence structural change in the genome induced by external factors. The accumulation of mutations eventually leads to cell phenotypic changes and even canceration (Negrini et al., 2010). Moreover, the application of radiotherapy in cancers typically relies on this accumulation of genomic instability (Huang & Zhou, 2020, 2021; Huang et al., 2020). There are two common types of genomic instability, including microsatellite instability (MIN) at the gene level and chromosomal instability (CIN) at the chromosomal level.
Microsatellites are simple tandem repeats with 1–6 nucleotides as a repeating unit in the genome with no more than 60 repetitions. The most common microsatellite sequence is a dimer of cytosine and adenine. There are tens of thousands of microsatellite loci in the human genome, which are evenly distributed on each chromosome, accounting for about 3% of the entire genome. Microsatellite sequences have a wide range of polymorphisms between individuals, but they maintain certain stability within individuals and can maintain genetic stability in offspring. Therefore, microsatellite sequences can be used as an important genetic marker (Velzen et al., 2020). Microsatellite instability refers to the phenomenon of changes in the microsatellite sequence caused by the insertion or deletion of microsatellite repeat units during DNA replication (Yan et al., 2021). Mainly due to DNA polymerase slippage, one or more bases in the repeat sequence are mismatched and microsatellite recombination, resulting in base pair deletion or insertion. This process can be recognized and repaired by the mismatch repair system, and when the mismatch repair function declines, it causes an increase in DNA replication errors, which leads to MIN (Boland & Goel, 2010; Guo et al., 2020, 2021).
Chromosomes are the material basis of cell inheritance. Chromosomal instability refers to changes in the number and structure of chromosomes that occur during mitosis, manifested as the gain or loss of entire chromosome copies or chromosomal fragments, heterozygous deletions, chromosomal translocations, rearrangements, chromosomal homochromatic regions due to gene amplification, and double microsomes (Geigl et al., 2008). Chromosomal instability can be caused by a variety of functional abnormalities, such as DNA damage response, cell cycle checkpoint disorders, chromosome missegregation caused by defective centrosome or spindle formation, and telomere dysfunction (Chang et al., 2012; Ding et al., 2011; Gronroos & López-García, 2018; Ye et al., 2013). Molecular cytogenetic studies have shown that most tumor cells, especially solid tumor cells, have non-random abnormalities in chromosomal segments during the development process, which manifest as changes in the number or structure of chromosomes (Hu et al., 2014; Nie et al., 2019). These changes are related to the amplification and inhibition of proto-oncogenes. Deletions of oncogenes are closely related (Bakhoum & Cantley, 2018; Fujimura et al., 2020; Ji et al., 2020; Sansregret et al., 2018).
Genomic instability has recently been suggested to be regulated by mechanical signaling. Tissue homeostasis is required to maintain functional integrity under stress (Dupont & Wickström, 2022). The main source of stress is mechanical force, which acts on cells, nuclei, and chromatin, but how the genome is protected from mechanical stress is unclear.
The extracellular mechanical microenvironment regulates genomic instability
Cell phenotypes are modulated by a variety of mechanical stimuli in the microenvironment. When external biomechanical stimuli change beyond the adaptive capacity of cells, cells can regulate chromosome organization and genome changes to cope with these changes in mechanical characteristics, resulting in transdifferentiation, diseases and even tumors. Therefore, it is not surprising that there is an interaction between the mechanical microenvironment and genomic instability.
Matrix metalloproteinases (MMPs) are a class of proteolytic enzymes containing active Ca2+, Zn2+ and other metal ions. In the ECM, MMPs can hydrolyze various substances such as collagen, fibronectin, and elastin that reflect the stiffness of the ECM. In addition, they can induce the dissociation of cell junctions and the degradation of ECM, breaking through the physical limitations of cells. It plays a role in biological processes such as tissue remodeling and growth, wound repair, and even embryogenesis. MMPs can affect multiple intracellular signaling pathways, dividing intracellular targets, thereby inducing mitotic abnormalities and genomic instability (Orlichenko & Radisky, 2008).
Mechanosensing receptors regulate genomic instability.
The primary site where cells come into direct contact with the mechanical microenvironment and transmit forces into the cell is the cell membrane. The sites in contact with the ECM or stiff surfaces usually form discrete multiprotein complexes centered on integrins, called focal adhesions (FAs), which are the main hubs for the interaction between cells and the ECM. In addition, in vivo, cells rarely exist in isolation, and cell adhesions centered on cadherin are formed between cells. These mechanosensing structures are able to sense and transmit mechanical signals in the extracellular microenvironment to the cytoskeleton. Here, we discuss the regulation of genomic instability by cellular ECM and cell adhesion.
The viability of an organism requires the faithful segregation of chromosomes within germ cells and somatic cells. One of the important features of tumorigenesis and development is genomic instability caused by chromosome segregation errors. Chromosomal segregation errors can lead to chromosomal damage and aneuploidy, which in turn contribute to enhanced cancer fitness. Studies have shown that integrins can indirectly affect chromosome segregation by regulating the attachment of chromosomes to the spindle (Knouse et al., 2018). Integrin-mediated polarity influences mitotic spindle orientation through the interaction of polarizing proteins LGN and NuMA with dynein to position stellate microtubules (Bergstralh et al., 2013; Lechler & Fuchs, 2005). Integrin ligase (ILK) is also associated with genomic instability. ILK is a threonine/serine protein kinase that plays an important role in focal adhesion mechano-signaling. Studies have shown that ILK is involved in centrosome and spindle assembly. ILK-overexpressed colorectal cancer cells prevent nocodazole-induced mitotic arrest, aggravated DNA damage and replication stress, increased micronuclei, and increased genomic instability, which may be related to ILK depolymerizing microtubules or dysregulation of the spindle assembly checkpoint (Chadla et al., 2021).
Cytoskeletal systems regulate genome instability
The cytoskeleton is a fibrous network structure composed of filamentous proteins and cross-linked proteins, and its types include actin microfilaments (F-actin), microtubules (MTs) and intermediate filaments (IF). It provides mechanical support for cells and controls their movement, shape, and tension homeostasis. Mechanical signals sensed at the plasma membrane can be transmitted directly to the nucleus through the cytoskeletal network, which in turn affects different nuclear processes, including replication, transcription, chromosome remodeling, and DNA repair, resulting in genomic instability. Here, we mainly discuss the regulation of genomic instability by different components of the cytoskeleton.
The actin cytoskeleton contributes to the maintenance of genome stability. Studies have shown that exoenzyme C3 transferase can induce actin filament damage by inhibiting Rho GTPase and make the MeWo cell line more sensitive to ultraviolet light radiation. Changes in key proteins in the DNA damage repair pathway led to DNA sensitization and increased mutation accumulation, greatly increasing the pressure of genomic instability (Magalhaes et al., 2020). The microtubule backbone is one of the key mechanisms for spindle assembly, mitosis, and the faithful segregation of genetic material, suggesting that microtubules are also key structures for maintaining genome integrity and fidelity (Knouse et al., 2018). MTs respond to mechanical stress. After mechanical loading, dividing cells show mitotic spindle alignment parallel to the applied force. Aberrant spindle microtubule expression, assembly, localization, and post-transcriptional modifications can increase chromosome segregation pressure, resulting in increased chromosomal instability. Intermediate filament structures exist in the nucleus, and this intermediate filament nucleoskeleton plays an important role in maintaining nuclear structure and mechanical stability, as well as genome function and integrity.
Nuclear mechanotransduction regulates genomic instability
The mechanical properties of the nucleus are determined by the complex structure of cytoskeletal filaments, nuclear layers, and chromatin (Uhler & Shivashankar, 2017). Mechanical signals sensed at the plasma membrane are transmitted directly to the nucleus through the cytoskeletal network, where reorganization of nuclear structure, deformation in the direction of tension, and chromatin reorganization soon occur, ultimately regulating chromosome organization and genome expression (Chang et al., 2018; Sun, et al., 2020; Tajik et al., 2016). For example, when cells attach to an isotropic or soft substrate, due to loss of actomyosin contractility, increased compression of the nucleus by microtubules, decreased levels of lamin A/C, and increased chromatin condensation, etc. Combined, the nucleus becomes relaxed, which in turn alters histone post-translational modifications and chromatin structure, and leads to differences in the accessibility of transcription factors to gene regulatory sites on DNA (Buxboim et al., 2014). Furthermore, the relative position of chromatin in the nucleus depends on the mechanical state of the nucleus (Maharana et al., 2016). The reduction of ECM can also lead to the separation of part of chromatin from the nuclear envelope, increasing telomeres and heterochromatin (Makhija et al., 2016). For example, fibroblasts attached to polarized substrates have flat ellipsoid nuclei with chromosomes preferentially aligned along the nuclear mechanical axis parallel to the nuclear attachment. Cells attached to isotropic substrates have more spherical nuclei and chromosomes are also preferentially aligned along the nuclear mechanical axis, but due to changes in nuclear shape, the axis is perpendicular to their attachment.
Discussion
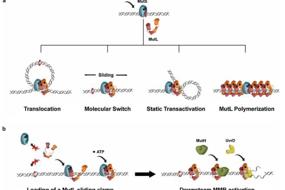
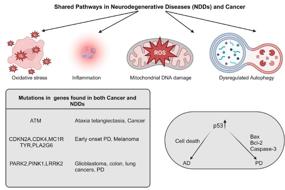
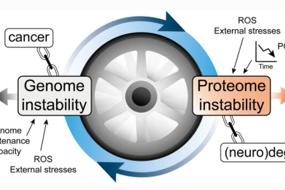
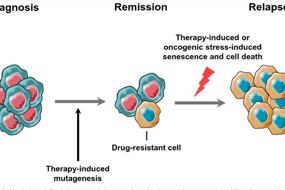
The idea that mechanical forces regulate cellular function touches several areas of research, including genomic instability. In recent years, more and more studies have shown that the abnormal mechanical microenvironment and the internal mechanical system of cells can affect the stability of chromosomes and genomes. Cells exposed to different mechanical environments have corresponded changes in shape and polarity, as well as different adhesion properties (Guo et al., 2021; Zheng et al., 2022). At the same time, disturbances in the mechanosensing receptors, cytoskeleton, and nuclear conduction systems within cells can prevent all aspects of chromosome segregation during cell division, forcing increased chromosomal instability. In this review, we provide a brief overview of how genomic instability is regulated by the tissue mechanical environment and subsequent mechanotransduction pathways (Fig. 1) .
Current research is not sufficient to comprehensively and systematically describe the interconnection between cell mechanics and genomic instability, and many questions remain to be addressed. For example, most studies so far have emphasized mechanotransduction related to cell-ECM and cell junctions, while studies on the effects of other types of mechanical stimuli, such as shear stress, compressive stress, and surface tension on genomic instability is less. In addition, current studies mostly focus on the regulatory mechanisms between mechanical signals and genomic instability caused by chromosomal abnormalities, and little has been discussed whether there is a physiological correlation between mechanical forces and microsatellite instability. To accomplish this, we need to use more sophisticated experimental instruments to measure changes in tissue and intracellular mechanics and genomes, and modulate them directly and specifically in vivo, and this is just the beginning.
The interconnectedness between cell mechanics and genomic instability highlight the necessity of mechanical forces as a key component of the cellular microenvironment and the importance of considering mechanical perturbations in culture conditions for in vitro studies of genomic instability. By introducing more physio-mechanical conditions and using existing or new mechano-detection techniques to study genomic instability, it may be possible to gain a better understanding of how genomic instability is regulated. These understandings can provide new perspectives on diseases caused by genomic instability, and also have important reference significance for the future development of drugs and methods targeting genomic instability and disease mechanical microenvironment.
Fig. 1
Schematic diagram about how mechanical factors affect genomic instability
References
Bakhoum, S. F., & Cantley, L. C. (2018). The multifaceted role of chromosomal instability in cancer and its microenvironment. Cell, 174(6), 1347–1360.
Bergstralh, D. T., Haack, T., & St Johnston, D. (2013). Epithelial polarity and spindle orientation: Intersecting pathways. Philosophical Transactions of the Royal Society B: Biological Sciences, 368(1629), 20130291.
Boland, C. R., & Goel, A. (2010). Microsatellite instability in colorectal cancer. Gastroenterology, 138(6), 2073-2087.e3.
Buxboim, A., et al. (2014). Matrix elasticity regulates lamin-A, C phosphorylation and turnover with feedback to actomyosin. Current Biology, 24(16), 1909–1917.
Chadla, P., et al. (2021). Integrin-linked-kinase overexpression is implicated in mechanisms of genomic instability in human colorectal cancer. Digestive Diseases and Sciences, 66(5), 1510–1523.
Chang, L., et al. (2012). miR-3928 activates ATR pathway by targeting Dicer. RNA Biology, 9(10), 1247–1254.
Chang, L., et al. (2018). The SWI/SNF complex is a mechanoregulated inhibitor of YAP and TAZ. Nature, 563(7730), 265–269.
Chen, X., & Song, E. (2022). The theory of tumor ecosystem. Cancer Communications (london), 42, 587–608.
Dieterle, M. P., et al. (2021). Integrins, cadherins and channels in cartilage mechanotransduction: Perspectives for future regeneration strategies. Expert Reviews in Molecular Medicine, 23, e14.
Ding, N., et al. (2011). Detection of novel human MiRNAs responding to X-ray irradiation. Journal of Radiation Research, 52(4), 425–432.
Dupont, S., & Wickström, S. A. (2022). Mechanical regulation of chromatin and transcription. Nature Reviews Genetics, 23, 624.
Fernandez-Sanchez, M. E., et al. (2015). Mechanotransduction’s impact on animal development, evolution, and tumorigenesis. Annual Review of Cell and Developmental Biology, 31, 373–397.
Fujimura, A., et al. (2020). Editorial: The role of epigenetic modifications in cancer progression. Frontiers in Oncology, 10, 617178.
Gao, S. S., et al. (2020). TIP60 K430 SUMOylation attenuates its interaction with DNA-PKcs in S-phase cells: Facilitating homologous recombination and emerging target for cancer therapy. Science Advances, 6(28), eaba7822.
Geigl, J. B., et al. (2008). Defining “chromosomal instability.” Trends in Genetics, 24(2), 64–69.
Gronroos, E., & López-García, C. (2018). Tolerance of chromosomal instability in cancer: mechanisms and therapeutic opportunities. Cancer Research, 78(23), 6529–6535.
Guo, Z., et al. (2020). HUWE1-dependent DNA-PKcs neddylation modulates its autophosphorylation in DNA damage response. Cell Death & Disease, 11(5), 400.
Guo, Z., et al. (2021). The long noncoding RNA CRYBG3 induces aneuploidy by interfering with spindle assembly checkpoint via direct binding with Bub3. Oncogene, 40(10), 1821–1835.
Hayward, M. K., Muncie, J. M., & Weaver, V. M. (2021). Tissue mechanics in stem cell fate, development, and cancer. Developmental Cell, 56(13), 1833–1847.
Hu, W., et al. (2014). MiR-663 inhibits radiation-induced bystander effects by targeting TGFB1 in a feedback mode. RNA Biology, 11(9), 1189–1198.
Huang, R., et al. (2020). BECN1 promotes radiation-induced G2/M arrest through regulation CDK1 activity: A potential role for autophagy in G2/M checkpoint. Cell Death Discov, 6, 70.
Huang, R. X., & Zhou, P. K. (2020). DNA damage response signaling pathways and targets for radiotherapy sensitization in cancer. Signal Transduction and Targeted Therapy, 5(1), 60.
Huang, R., & Zhou, P. K. (2021). DNA damage repair: Historical perspectives, mechanistic pathways and clinical translation for targeted cancer therapy. Signal Transduction and Targeted Therapy, 6(1), 254.
Humphrey, J. D., Dufresne, E. R., & Schwartz, M. A. (2014). Mechanotransduction and extracellular matrix homeostasis. Nature Reviews Molecular Cell Biology, 15(12), 802–812.
Jeggo, P. A., Pearl, L. H., & Carr, A. M. (2016). DNA repair, genome stability and cancer: A historical perspective. Nature Reviews Cancer, 16(1), 35–42.
Ji, M. T., et al. (2020). 1α,25(OH)(2)D(3) Radiosensitizes cancer cells by activating the NADPH/ROS pathway. Frontiers in Pharmacology, 11, 945.
Jin, P., Jan, L. Y., & Jan, Y. N. (2020). mechanosensitive ion channels: Structural features relevant to mechanotransduction mechanisms. Annual Review of Neuroscience, 43, 207–229.
Kefauver, J. M., Ward, A. B., & Patapoutian, A. (2020). Discoveries in structure and physiology of mechanically activated ion channels. Nature, 587(7835), 567–576.
Knouse, K. A., et al. (2018). Chromosome segregation fidelity in epithelia requires tissue architecture. Cell, 175(1), 200-211.e13.
Lechler, T., & Fuchs, E. (2005). Asymmetric cell divisions promote stratification and differentiation of mammalian skin. Nature, 437(7056), 275–280.
Magalhaes, Y. T., Cardella, G. D., & Forti, F. L. (2020). Exoenzyme C3 transferase lowers actin cytoskeleton dynamics, genomic stability and survival of malignant melanoma cells under UV-light stress. Journal of Photochemistry and Photobiology b: Biology, 209, 111947.
Maharana, S., et al. (2016). Chromosome intermingling—The physical basis of chromosome organization in differentiated cells. Nucleic Acids Research, 44(11), 5148–5160.
Makhija, E., Jokhun, D. S., & Shivashankar, G. V. (2016). Nuclear deformability and telomere dynamics are regulated by cell geometric constraints. Proceedings of the National Academy of Sciences of the United States of America, 113(1), E32-40.
Negrini, S., Gorgoulis, V. G., & Halazonetis, T. D. (2010). Genomic instability—An evolving hallmark of cancer. Nature Reviews Molecular Cell Biology, 11(3), 220–228.
Nie, Y. H., et al. (2019). Analysis of mRNA expression patterns in peripheral blood cells of 3 patients with cancer after the first fraction of 2 Gy irradiation: an integrated case report and systematic review. Dose Response, 17(1), 1559325819833474.
Orlichenko, L. S., & Radisky, D. C. (2008). Matrix metalloproteinases stimulate epithelial-mesenchymal transition during tumor development. Clinical & Experimental Metastasis, 25(6), 593–600.
Sansregret, L., Vanhaesebroeck, B., & Swanton, C. (2018). Determinants and clinical implications of chromosomal instability in cancer. Nature Reviews. Clinical Oncology, 15(3), 139–150.
Schreiber, T. H., et al. (2007). Shear flow-dependent integration of apical and subendothelial chemokines in T-cell transmigration: Implications for locomotion and the multistep paradigm. Blood, 109(4), 1381–1386.
Strzyz, P. (2016). Mechanotransduction: May the force be with you. Nature Reviews Molecular Cell Biology, 17(9), 533.
Sun, J., et al. (2020). Force-induced gene up-regulation does not follow the weak power law but depends on H3K9 demethylation. Science Advances, 6(14), eaay9095.
Sun, Z., Guo, S. S., & Fässler, R. (2016). Integrin-mediated mechanotransduction. Journal of Cell Biology, 215(4), 445–456.
Tajik, A., et al. (2016). Transcription upregulation via force-induced direct stretching of chromatin. Nature Materials, 15(12), 1287–1296.
Uhler, C., & Shivashankar, G. V. (2017). Regulation of genome organization and gene expression by nuclear mechanotransduction. Nature Reviews Molecular Cell Biology, 18(12), 717–727.
van Velzen, M. J. M., et al. (2020). MSI as a predictive factor for treatment outcome of gastroesophageal adenocarcinoma. Cancer Treatment Reviews, 86, 102024.
Wozniak, M. A., & Chen, C. S. (2009). Mechanotransduction in development: A growing role for contractility. Nature Reviews Molecular Cell Biology, 10(1), 34–43.
Yan, S., et al. (2021). ZGRF1 promotes end resection of DNA homologous recombination via forming complex with BRCA1/EXO1. Cell Death Discov, 7(1), 260.
Ye, C., et al. (2013). Radiation-induced cellular senescence results from a slippage of long-term G2 arrested cells into G1 phase. Cell Cycle, 12(9), 1424–1432.
Zheng, L., et al. (2022). Ionizing radiation-induced long noncoding RNA CRYBG3 regulates YAP/TAZ through mechanotransduction. Cell Death & Disease, 13(3), 209.
Acknowledgements
This work is supported by National Natural Science Foundation of China (Grants No: 31971165 and 82173465); The Fok Ying-Tong Education Foundation, China (Grant No: 171017); The Natural Science Foundation of Jiangsu Province (Grant No: KB20191422); Leading Talents Program of Gusu District, Suzhou City (Grant No: ZXL2022454).
Author information
Authors and Affiliations
State Key Laboratory of Radiation Medicine and Protection, School of Radiation Medicine and Protection, Collaborative Innovation Center of Radiation Medicine of Jiangsu Higher Education Institutions, Medical College of Soochow University, Suzhou, China
Lijun Zheng, Mintao Ji & Lei Chang
The First Affiliated Hospital of Soochow University, Suzhou, China
Hong Zhang
Corresponding author
Correspondence to Lei Chang.
Ethics declarations
Conflict of interest
Authors declare that they have no competing interests.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zheng, L., Ji, M., Zhang, H. et al. Regulatory mechanisms of mechanotransduction in genome instability. GENOME INSTAB. DIS. 3, 311–316 (2022). //doi.org/10.1007/s42764-022-00086-x
Received19 July 2022
Revised21 September 2022
Accepted28 September 2022
Published25 October 2022
Issue DateDecember 2022
DOI//doi.org/10.1007/s42764-022-00086-x
Share this article
Anyone you share the following link with will be able to read this content:
Get shareable linkKeywords
Genome instability
Mechanotransduction

用户登录
还没有账号?
立即注册